About
I’m a scientist and engineer who thrives at the intersection of science, software, and hardware.
I was a Postdoctoral Scholar in the Moore Lab at the Scripps Institution of Oceanography and UC San Diego. I joined the Moore Lab in July 2019, and was a self-funded NIH/NIEHS F32 Postdoctoral Fellow in the Moore lab from 2020 through 2023. I completed my PhD in the Weng Laboratory of the Department of Biology of the Massachusetts Institute of Technology.
My postdoctoral research elucidated the first known causal biosynthetic genes (the “PKZILLAs”) for an algal giant marine polyether toxin - an impactful class of molecules of significant pharmaceutical and toxicological interest.
One of the PKZILLAs also turned out to be larger than titin, the accepted largest known protein: 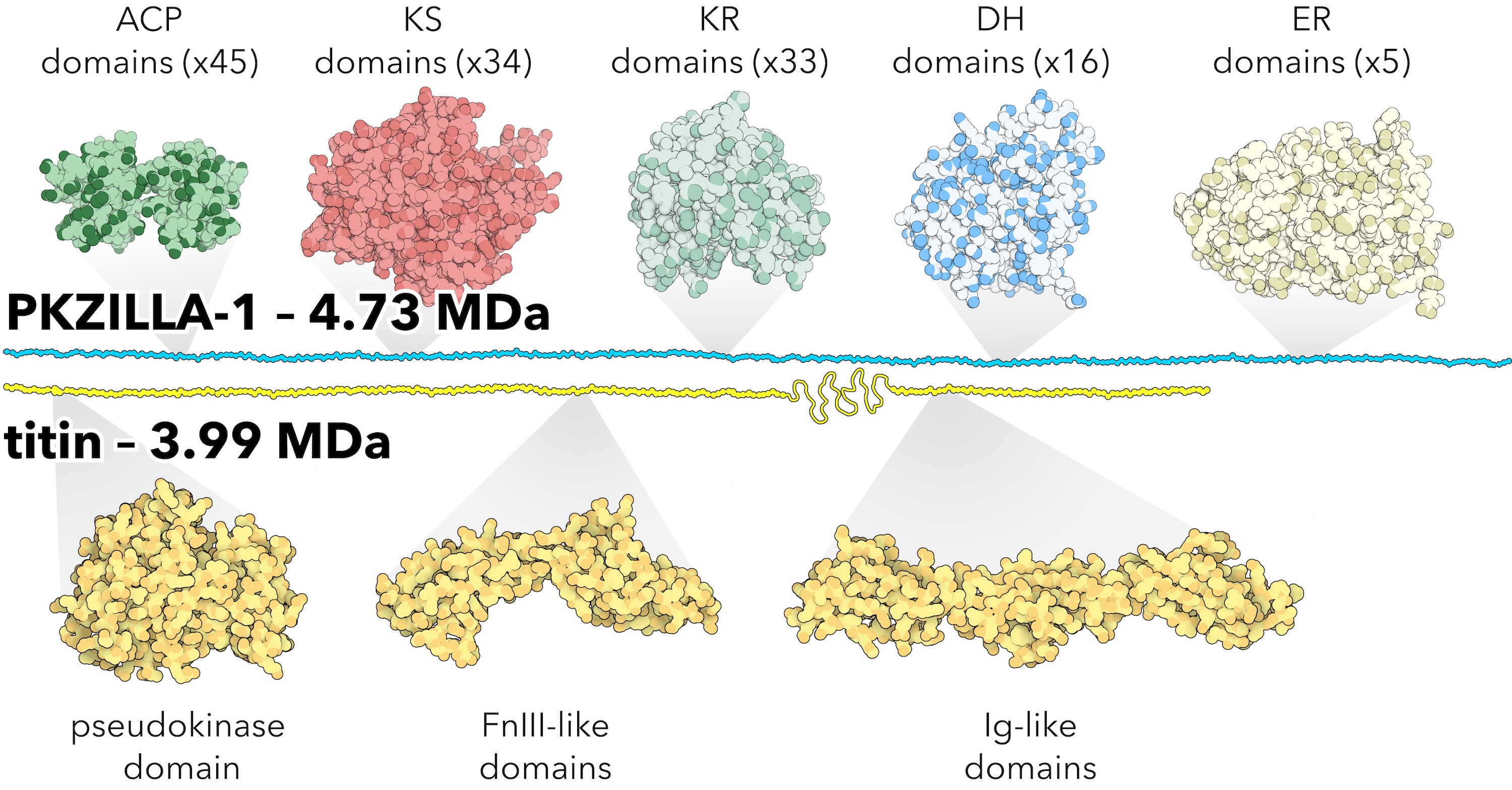 Figure by Tim Fallon, PhD. Stylistic size comparison of PKZILLA-1 and human titin, not to scale, wherein PKZILLA-1 is shown as roughly 25% larger than titin. Abbreviations: megadalton (MDa), acyl carrier protein (ACP), ketosynthase (KS), ketoreductase (KR), dehydratase (DH), enoylreductase (ER), fibronectin (Fn), immunoglobulin (Ig). Titin figure adapted from https://pdb101.rcsb.org/motm/185 under Creative Commons license, attribution: David S. Goodsell, RCSB Protein Data Bank (PDB). ACP structure modeled from bacterial ACP (PDB id: 2FAE). KS, KR, DH, ER, modeled from mammalian fatty acid synthase (PDB id: 2VZ8).
Figure by Tim Fallon, PhD. Stylistic size comparison of PKZILLA-1 and human titin, not to scale, wherein PKZILLA-1 is shown as roughly 25% larger than titin. Abbreviations: megadalton (MDa), acyl carrier protein (ACP), ketosynthase (KS), ketoreductase (KR), dehydratase (DH), enoylreductase (ER), fibronectin (Fn), immunoglobulin (Ig). Titin figure adapted from https://pdb101.rcsb.org/motm/185 under Creative Commons license, attribution: David S. Goodsell, RCSB Protein Data Bank (PDB). ACP structure modeled from bacterial ACP (PDB id: 2FAE). KS, KR, DH, ER, modeled from mammalian fatty acid synthase (PDB id: 2VZ8).
My PhD research focused on characterizing the specialzed metabolism of firefly luciferin, and involved de novo transcriptomic and genomic sequencing and assembly, recombinant and crude enzymology, and LC-MS. I was a founding member of the Photinus pyralis (Big Dipper firefly) genome project, leading to the first linkage group resolution genome of any firefly. I’m interested in comparative genomic approaches to decipher metabolic questions generally.
Key findings of my PhD research include:
- Sequencing and assembly of the Photinus pyralis (Big Dipper Firefly) genome, and its comparison to other bioluminescent beetles, leading to the finding that click-beetle and firefly luciferase evolved separately
- The discovery of sulfoluciferin and luciferin sulfotransferase, which we now understand are conserved in all fireflies (Lampyridae)
More information
- LinkedIn - timfallonphd
- GitHub - photocyte
- GitLab - photocyte
- X.com - @photocyte@x.com
- Mastodon - @photocyte@genomic.social
- Academic style Curriculum vitae (CV): Timothy_R_Fallon_CV_PW-is-Spring2022.pdf (CV password protected with Spring2022 to prevent webcrawler email spam)
- ORCID: 0000-0002-3048-7679
- Google Scholar publications and citations
Miscellaneous endevours
- In 2017 We launched the datasite for the firefly genome project: http://www.fireflybase.org (Now sunsetted, and data migrated to NCBI)
- In 2016 Crowdfunded the project to sequence the Photinus pyralis genome on the Experiment.com Scientific crowdfunding platform: https://experiment.com/projects/illuminating-the-firefly-genome
- In 2014 I took the “How to Make Almost Anything” class at the MIT media lab. You can see my exploits here: http://fab.cba.mit.edu/classes/863.14/people/tim_fallon/index.html
Academic Collaborators
- Wisecaver lab (WSU) https://labs.wsu.edu/wisecaver/
- Moore Lab (SIO/UCSD) http://scrippsscholars.ucsd.edu/bsmoore
- Weng Lab (MIT): https://wenglab.net
- Lewis Lab (Tufts): https://ase.tufts.edu/biology/faculty/lewis/
- Sarah Lower (Bucknell University): https://thelowerlab.wordpress.com
- Bybee lab (BYU): http://bybeelab.byu.edu/Home.aspx
- Stanger-Hall lab (UGA): http://research.franklin.uga.edu/stanger-hall/
- Oba lab (Chubu University): https://www3.chubu.ac.jp/faculty/oba_yuichi/english_page/
- Fireflyers International: https://fireflyersinternational.net/